A Tensor SVD-based Classification Algorithm Applied to fMRI Data
This post was written by Katy Keegan, Yihua Xu, Tanvi Vishwanath, and Vida Jon and published with minor edits. The team was advised by Dr. Elizabeth Newman. In addition to this post, the team has also given a midterm presentation , created a poster , published, code, and written a paper.
Overview: Can we look at a brain scan and know what the brain’s owner is thinking?
Our research attempts to use computers to correctly classify brain scans into 2 groups, depending on what they are doing during the scan.
Functional MRI
We use brain scans called functional MRIs that show us which parts of the brain are using more oxygen and are therefore most active. We have fMRIs of test subjects who, as they are being scanned, are also shown either a picture or a sentence. If computers can classify these study subjects into one of these two categories by only studying their scans, then in a sense we can read their minds.
fMRIs consist of three-dimensional pixels called voxels, and the data are numbers representing colors. Unlike static MRIs which take a scan at one point in time, fMRIs are repeated every few seconds creating a series of images for each trial. Here is an example of an fMRI of one brain during one trial. The different images are of different slices of the brain. The abbreviations on the right refer to brain regions, for example “SMA” stands for “Supplementary Motor Area” located at the top center of the head.

If we have three-dimensional fMRI brain voxel data for many patients, multiple scans in sequence, then we need to analyze a quantity of data unwieldy even for modern computers.
Image Classification
Image classification is using a computer to figure what what an image represents. Computers can’t see images, so they use features of the images that it can understand. For example, we can train a computer to match an image to a numerical digit. Computers learn by training on many images, for example using the MNIST database of handwritten images. MNIST contains a wide variety of images that can represent a 0 or a 1:
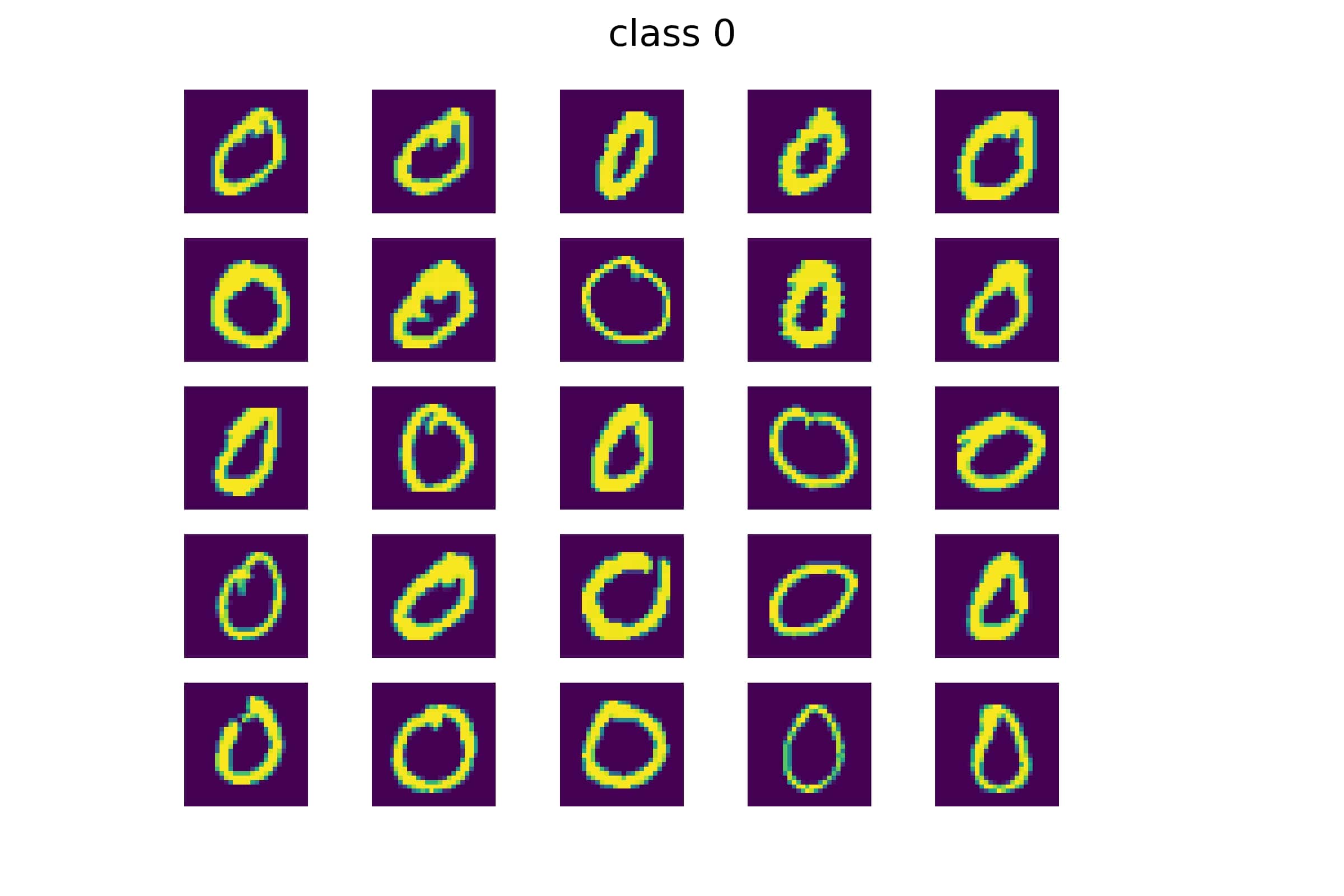

We can construct a “basis”, which can be thought of as a collection of the most relevant features shared by all of the images belonging to that class.
We see that the basis for Class 0 shows more curved features, while the basis for Class 1 contains traces of more straight and vertical features. To choose the basis the image more closely matches, we compute a “projection”, not unlike what you may have calculated with vectors. The larger the projection, the better the match to a particular basis.

The computer tries to classify this test image as either a zero or a one:

This image represents the projection of the test image onto the basis for numeral one:

The image projection onto the basis for zero, shows differences and inconsistencies:

The projection shows that the “distance” between our test image and our two classes is smaller for Class 1 than for Class 0, and so our method classifies our test image as a 1.
By learning from a training set of images a computer can examine the data in a new image and figure out which digit it most resembles. Similarly, we are training our computer to learn how to use the data in an fMRI to classify our study subjects into those who are shown an image and those who are shown a sentence.
Tensors and Singular Value Decomposition
Typically, large data sets like the fMRI voxels are stored in matrices, which have some powerful tools for extracting the most relevant components.
When we store fMRI data in a matrix we lose important relationships between the data points. For example a computer does not know that a particular voxel representing a part of the brain at one moment is that same part of the brain a few seconds later.
In our work, we study how to store our data in a tensor which is like matrix but with more than 2 dimensions. Our tensor of fMRIs have a total of 5 dimensions, shown in the figure below. The green slices consist of voxels of the brain in 3 spatial dimensions: x,y,z (yellow). For each trial multiple images are taken over several seconds (blue), and there are multiple trials (red).
<img width=“600” alt=“fmri_tensors”
When we store our voxel data in tensors instead of matrices, we retain all the information about the 3 dimensional location in the brain, the sequence of the image in time, and the trial. Now we want to decompose our tensor data using a method analogous to the way matrices can be decomposed, so we can use far less data and still extract an accurate prediction of what the subject is thinking. In matrices this is called Singular Value Decomposition; we call our approach tensor-SVD or tSVD.
Here is a diagram of matrix decomposition using SVD:

This what we imagine a tensor SVD would be:

You’ll notice that we use matrix multiplication in working with matrix SVDs. We are searching for the equivalent tensor multiplication.
Other Applications
Our research is not only useful for fMRIs; many datasets have multiple dimensions. Streaming entertaining companies have data on thousands of viewers and what movies they’ve watched. Hospitals track thousands of patients, each of whom has had multiple lab tests and other studies. If our research enables us to classify our fMRI subjects, then we may also be able to predict whether someone will want to watch Terminator, or whether a patient is likely to have cancer.
References
Tensor tensor products with invertible linear transforms
Tensor-tensor algebra for optimal representation and compression of multiway data
Tensor decompositions and applications
Image classification using local tensor singular value decompositions